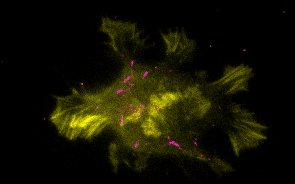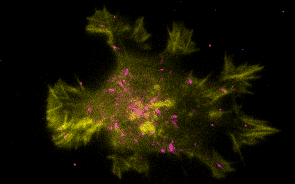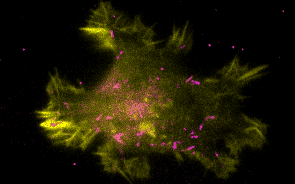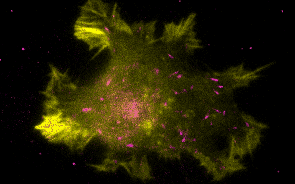
Research Overview
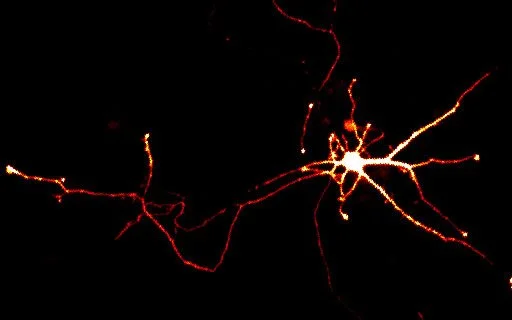
Research in the Goode lab focuses on defining the biochemical and cellular mechanisms of actin and microtubule cytoskeleton rearrangements that drive cell morphogenesis, motility, endocytosis, and intracellular transport. To tackle these problems, we take a multi-disciplinary ‘top down’ and ‘bottom up’ approach, combining forward and reverse genetics, biochemistry, structural biology, cell imaging, and multi-wavelength single molecule TIRF microscopy. From 2000-2010, our primary in vivo discovery system was the budding yeast S. cerevisiae; however, since then we have made a major transition, and about half of the lab currently studies cytoskeletal dynamics in mammalian cells.
Our long-range goal is to define in molecular detail the multi-component mechanisms that underlie these critical steps in cytoskeletal remodeling, which govern cell shape, organization, and dynamics.
Our research interests:
• Coordination of microtubule and actin polymer dynamics
• Actin filament disassembly & remodeling
• Cellular actin assembly mechanisms
Microtubule-Actin Cross-Regulation
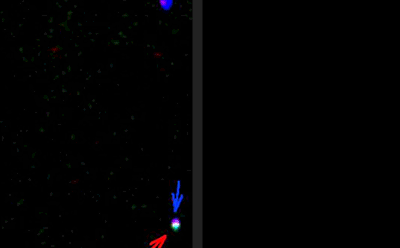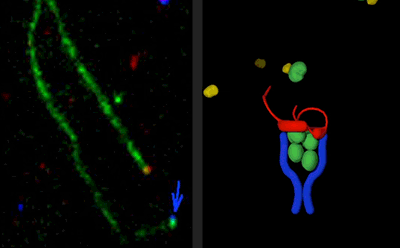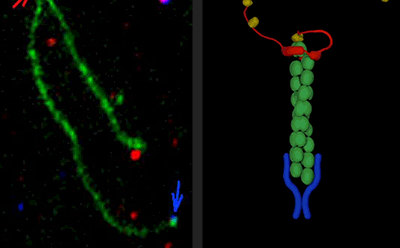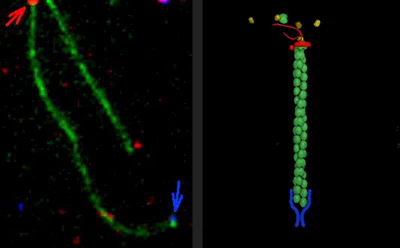
Historically, the microtubule and actin cytoskeletons have been studied separately. However, ultrastructural, pharmacological, and genetic evidence dating back decades has suggested that tight coordination between the microtubule and actin polymer networks is required for such fundamental processes as directed cell migration, cell division, and phagocytosis. In many of these biological settings, the growth of microtubule plus-ends into actin-rich cortical regions triggers changes in actin assembly-based functions, including focal adhesion turnover, filopodial extension, lamellipodia protrusion, neurite outgrowth, and phagocytic cup formation. We are studying the molecular mechanisms of microtubule-actin crosstalk underlying these processes, with particular focus on the roles of CLIP-170, formins, APC, EB1, IQGAP, and more.
Actin Disassembly and Remodeling
We have great interest in how cellular actin structures vary in their sizes, shapes, and turnover dynamics. The dynamics of each structure appear to be tailored to their biological functions. We are studying how this is achieved by cells using a core set of conserved actin disassembly-promoting factors including Cofilin, Coronin, Aip1, Srv2/CAP, Abp1, GMF, and Twinfilin. Recent results from our lab have demonstrated that many of these factors combine to work in complex multi-component mechanisms. This has included mechanisms of filament debranching, enhanced severing, and filament end-depolymerization, and how these mechanisms drive the rearrangement of entire actin filament networks with different geometries.
Cellular Actin Assembly Mechanisms
Cells rely on the precisely tuned activities of multiple actin assembly-promoting factors to generate diverse actin structures. These include the Arp2/3 complex, Formins, and APC. We are studying how the potent activities of these actin nucleation and elongation factors are spatially and temporary controlled in cells through interactions with a variety of binding partners. For the Arp2/3 complex, this includes WASp, WAVE, Coronin, GMF, and Abp1. For Formins, this includes Bud6, Bud14, Smy1, Hof1, APC, CLIP-170, Spire, and Capping Protein. We are studying these mechanisms by multi-wavelength single-molecule analysis in vitro and by complimentary in vivo experiments using genetics and live-cell imaging.